
Mapping Vector Data in R
Tufts Data Lab Workshop Spring 2017
Aishwarya Venkat
Data Lab Assistant
Welcome!
Expected previous knowledge:
RStudio basics (data frames, installing packages)
GIS basics (projections, attribute tables)
Why use R to make maps?
Automation/loops for downloading multiple datasets
More control over styles and symbology
Streamlined workflow (sometimes)
Outline
Review spatial data structures
Download shapefile from web, map world population
Map access to water around the world using World Bank's World Development Indicators (WDI) database
Map 2012 state population using choroplethr package
Make county-level choropleth map of household income in USA using American Community Survey (ACS) data
Goals
Become familiar with spatial data structures and sources
Translate geospatial methods (projections, symbology, legends, attributes) to R
Make pretty and informative maps :)
Step 1: Get set up
- Install packages:
packages<-c("choroplethr", "choroplethrMaps", "acs", "WDI", "gstat")
install.packages(packages)
- We will also use pre-installed packages sp, rgdal, dplyr, and ggplot2 during this session
packages<-c("choroplethr", "choroplethrMaps", "acs", "WDI", "sp", "rgdal",
"ggplot2", "gstat", "dplyr")
lapply(packages, require, character.only = TRUE)
Overview of Spatial Data Structures in R
- Data frames are similar to attribute tables in GIS
Overview of Spatial Data Structures in R
Each feature in attribute table is associated with discrete spatial information (point, line, or polygon)
Vector data = points, lines, polygons (package: sp)
- Points: SpatialPoints, SpatialPointsDataFrame
- Lines: SpatialLines, SpatialLinesDataFrame
- Polygons: SpatialPolygons, SpatialPolygonsDataFrame
Raster data = pixel/gridded data (packages: rgdal, raster)
- Rasters, raster stacks/bricks, etc.
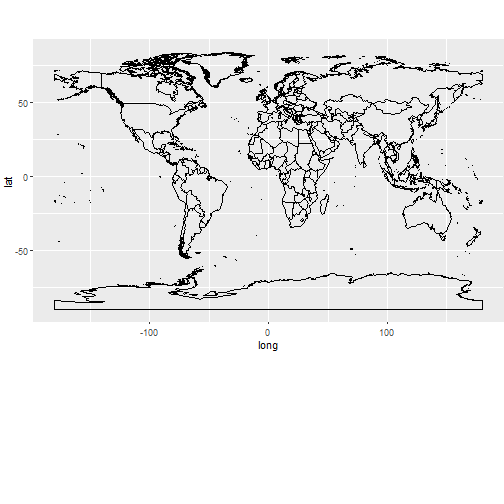
Polygon data: World
Download and map world borders
download.file("http://thematicmapping.org/downloads/TM_WORLD_BORDERS_SIMPL-0.3.zip",
destfile="world.zip"); unzip("world.zip");
worldmap=readOGR(dsn= getwd(), layer="TM_WORLD_BORDERS_SIMPL-0.3")
ggplot(worldmap, aes(x = long, y = lat, group = group)) + geom_path()+
theme(aspect.ratio=0.6, plot.margin = unit(c(0, 0, 4, 0), "cm"))
Explore worldmap
- What kind of object is it? What projection is it in? What data does it contain?
str(worldmap)
worldmap@proj4string
str(worldmap@data)
head(worldmap@data)
## FIPS ISO2 ISO3 UN NAME AREA POP2005 REGION SUBREGION
## 0 AC AG ATG 28 Antigua and Barbuda 44 83039 19 29
## 1 AG DZ DZA 12 Algeria 238174 32854159 2 15
## 2 AJ AZ AZE 31 Azerbaijan 8260 8352021 142 145
## 3 AL AL ALB 8 Albania 2740 3153731 150 39
## 4 AM AM ARM 51 Armenia 2820 3017661 142 145
## 5 AO AO AGO 24 Angola 124670 16095214 2 17
## LON LAT
## 0 -61.783 17.078
## 1 2.632 28.163
## 2 47.395 40.430
## 3 20.068 41.143
## 4 44.563 40.534
## 5 17.544 -12.296
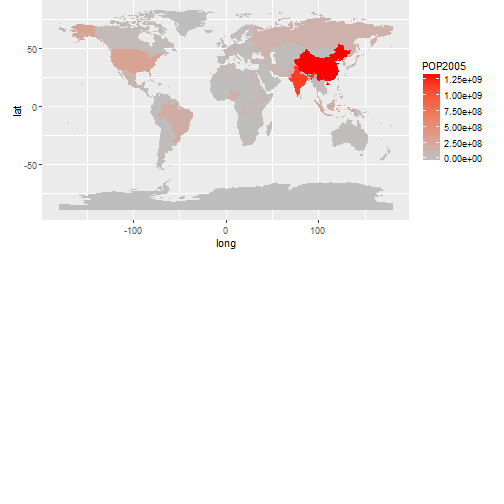
Map 2005 population with ggplot
library(RColorBrewer); library(maptools);
worldmap@data$id=rownames(worldmap@data)
worldmap_points<-fortify(worldmap, region="id")
worldmap_df<-join(worldmap_points, worldmap@data, by="id")
ggplot(worldmap_df, aes(long, lat, group=group, fill=POP2005))+geom_polygon()+
scale_fill_continuous(low="grey", high="red")+ coord_fixed() +
theme(aspect.ratio=0.6, plot.margin = unit(c(0, 0, 9, 0), "cm"))
World Development Indicators
Create world map of 2010 Access to water
library(WDI); library(rworldmap);
datamap = WDI(indicator = "SH.H2O.SAFE.ZS", start=2010, end=2010);
SPDF <- joinCountryData2Map(datamap, joinCode = "ISO2", nameJoinColumn = "iso2c");
mapParams <- mapCountryData(SPDF, nameColumnToPlot='SH.H2O.SAFE.ZS', mapTitle="",
mapRegion="world", catMethod="quantiles", numCats=10, colourPalette='heat');
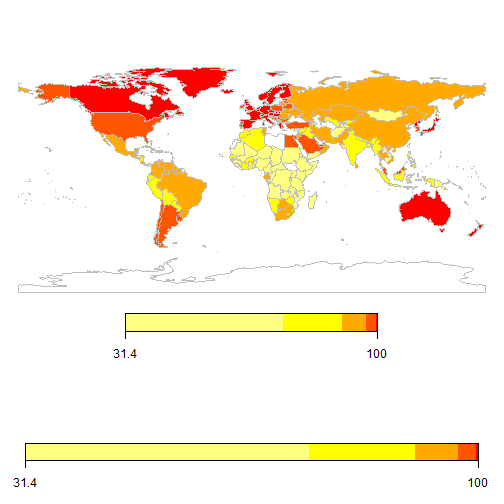
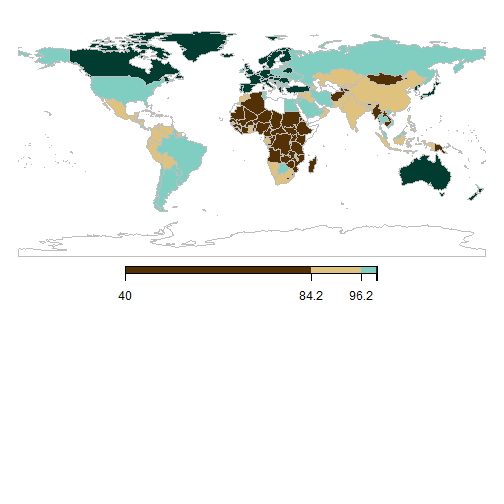
Changing legends and using RColorBrewer scales
cp <- brewer.pal(10,'BrBG')
datamap = WDI(indicator = "SH.H2O.SAFE.ZS", start=2014, end=2014);
SPDF <- joinCountryData2Map(datamap, joinCode = "ISO2", nameJoinColumn = "iso2c");
mapParams <- mapCountryData(SPDF, nameColumnToPlot='SH.H2O.SAFE.ZS', addLegend=TRUE,
mapRegion="world", catMethod="quantiles", numCats=10, colourPalette=cp, mapTitle = "")
do.call(addMapLegend, c(mapParams,legendLabels="all",legendWidth=0.5,legendIntervals="data",
legendShrink=0.5, legendMar = 16))
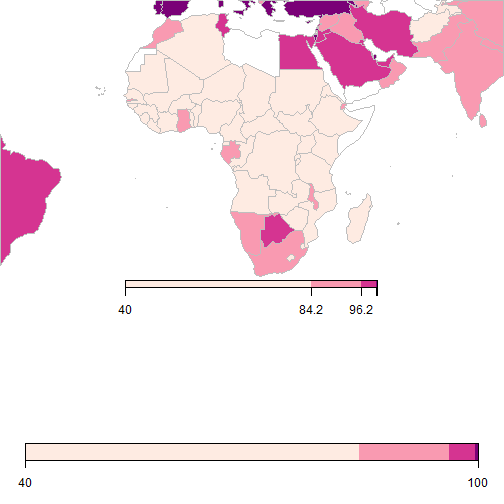
Zooming into Africa
cp <- brewer.pal(5,'RdPu'); datamap = WDI(indicator = "SH.H2O.SAFE.ZS", start=2014, end=2014)
SPDF <- joinCountryData2Map(datamap, joinCode = "ISO2", nameJoinColumn = "iso2c")
mapParams <- mapCountryData(SPDF, nameColumnToPlot='SH.H2O.SAFE.ZS', addLegend=TRUE,
mapRegion="Africa", catMethod="quantiles", numCats=10, colourPalette=cp)
do.call(addMapLegend, c(mapParams,legendLabels="all",legendWidth=0.5,legendIntervals="data",
legendShrink=0.5, legendMar = 15))
American Community Survey data
Load and Explore choroplethR data
library(choroplethr)
library(choroplethrMaps)
data(state.regions)
head(state.regions)
## region abb fips.numeric fips.character
## 1 alaska AK 2 02
## 2 alabama AL 1 01
## 3 arkansas AR 5 05
## 4 arizona AZ 4 04
## 5 california CA 6 06
## 6 colorado CO 8 08
data(df_pop_state)
head(df_pop_state)
## region value
## 1 alabama 4777326
## 2 alaska 711139
## 3 arizona 6410979
## 4 arkansas 2916372
## 5 california 37325068
## 6 colorado 5042853
State map of population
state_choropleth(df_pop_state, title = "2012 Population Estimates",
legend = "Population", buckets=3)
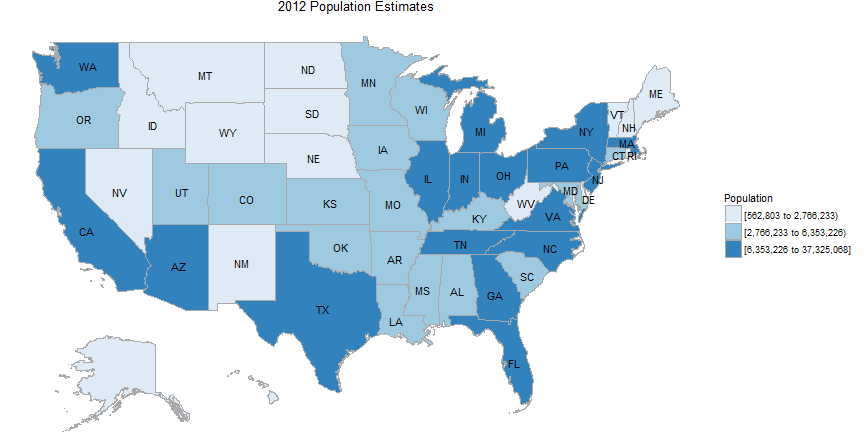
State map of population
state_choropleth(df_pop_state, title = "2012 Population Estimates", legend = "Population",
buckets=3, zoom=c("connecticut", "maine", "massachusetts", "new hampshire", "vermont",
"rhode island", "new jersey", "new york", "pennsylvania"))
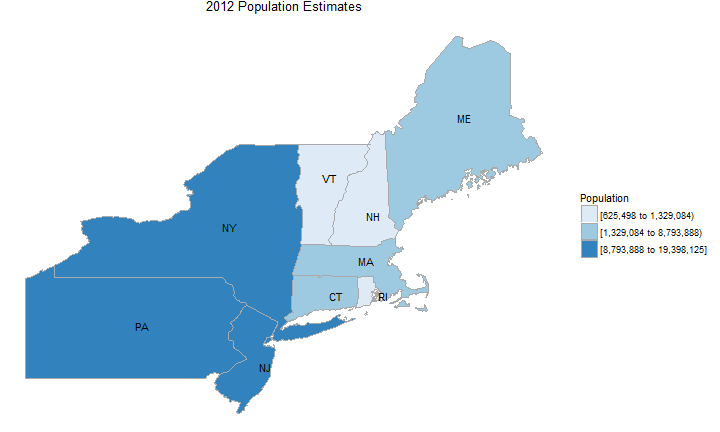
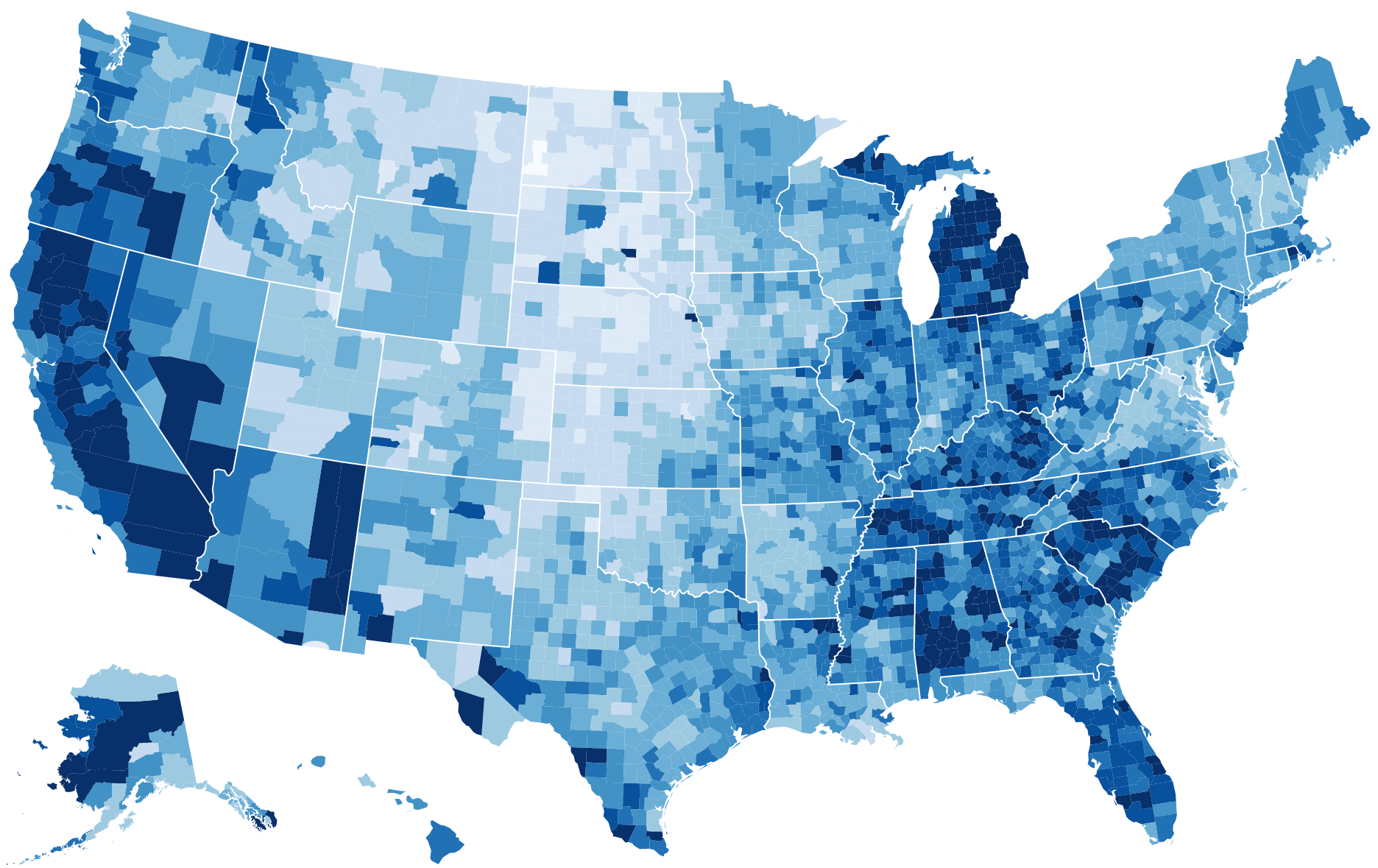
County map of county demographics
data("df_county_demographics")
df_county_demographics$value = df_county_demographics$percent_hispanic
county_choropleth(df_county_demographics, title="Percent Hispanic Residents (2012)", num_colors=5)
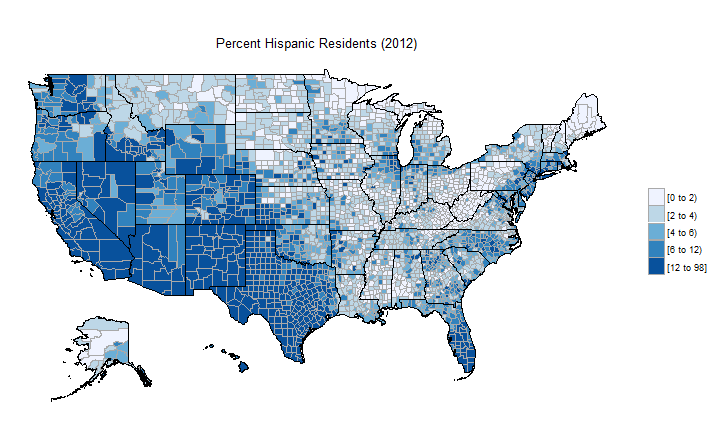
Downloading county-level ACS data
library(acs); api.key.install(key=""); par(mar=c(0,0,0,0), mai=c(0,0,0,0))
county_choropleth_acs("B19301", endyear=2014, span=5, num_colors=5)
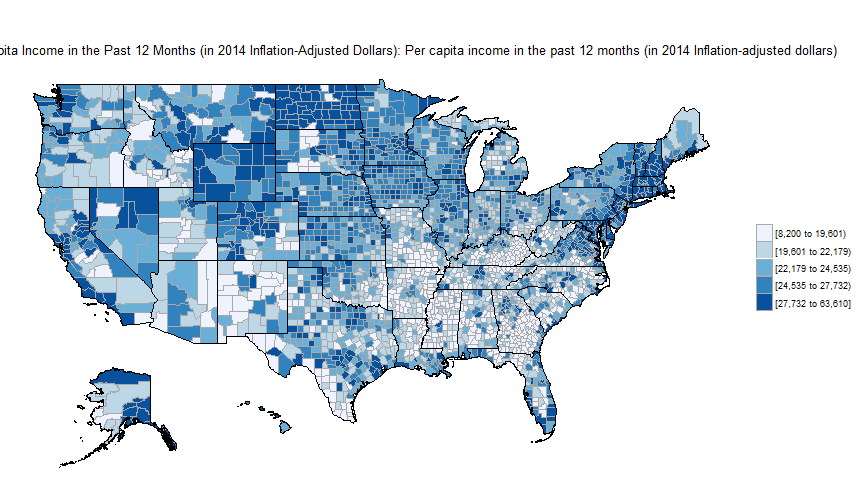
Exercises
Exercise 1: Use American Fact Finder to find and map a census variable
query<-acs.lookup(2013, span = 5, dataset = "acs", table.name = "income", case.sensitive=F)
#head(unique(query@results$table.number))
county_choropleth_acs("B01001D", endyear=2013, span=5, num_colors=5)
Exercise 2: Use WDI dataset to map one indicator for a region of your choice
WDIsearch('water'); water = WDI(indicator='SH.H2O.SAFE.ZS', country="all", start=2014, end=2014);
water <- subset(water, !is.na(water$SH.H2O.SAFE.ZS));
water <- group_by(water, country)
water <- filter(water, year == max(year))
names(worldmap_df)[9] <- "iso_a2"
final <- left_join(worldmap_df, water, by="iso_a2")
# Refer to this list for UN regions/subregions designations:
# http://unstats.un.org/unsd/methods/m49/m49regin.htm
# final <- subset(final, REGION==142)
ggplot(final) +
geom_path(aes(long, lat, group = group), color = "white") +
geom_polygon(aes(long, lat, group = group, fill = SH.H2O.SAFE.ZS)) +
scale_fill_gradient("% access to\nimproved water\nsource", low = "#132B43",
high = "#56B1F7", na.value = "grey50")
References
Ezra Glenn, Using acs.R to create choropleth maps
Ari Lamstein, Analyzing Census Data in R
Trulia Blog, The choroplethr package for R
Andrew Bonneau, WDI + rworldmap for world map data plotting in Rstudio
Ramnath Vaidyanathan, Slidify presentations from RMarkdown
Advanced examples
Demo dataset: meuse
meuse dataset: concentration of four heavy metals measured in the top soil in a flood plain along the river Meuse, with additional variables
library(sp);
data(meuse); summary(meuse); str(meuse);
## x y cadmium copper
## Min. :178605 Min. :329714 Min. : 0.200 Min. : 14.00
## 1st Qu.:179371 1st Qu.:330762 1st Qu.: 0.800 1st Qu.: 23.00
## Median :179991 Median :331633 Median : 2.100 Median : 31.00
## Mean :180005 Mean :331635 Mean : 3.246 Mean : 40.32
## 3rd Qu.:180630 3rd Qu.:332463 3rd Qu.: 3.850 3rd Qu.: 49.50
## Max. :181390 Max. :333611 Max. :18.100 Max. :128.00
##
## lead zinc elev dist
## Min. : 37.0 Min. : 113.0 Min. : 5.180 Min. :0.00000
## 1st Qu.: 72.5 1st Qu.: 198.0 1st Qu.: 7.546 1st Qu.:0.07569
## Median :123.0 Median : 326.0 Median : 8.180 Median :0.21184
## Mean :153.4 Mean : 469.7 Mean : 8.165 Mean :0.24002
## 3rd Qu.:207.0 3rd Qu.: 674.5 3rd Qu.: 8.955 3rd Qu.:0.36407
## Max. :654.0 Max. :1839.0 Max. :10.520 Max. :0.88039
##
## om ffreq soil lime landuse dist.m
## Min. : 1.000 1:84 1:97 0:111 W :50 Min. : 10.0
## 1st Qu.: 5.300 2:48 2:46 1: 44 Ah :39 1st Qu.: 80.0
## Median : 6.900 3:23 3:12 Am :22 Median : 270.0
## Mean : 7.478 Fw :10 Mean : 290.3
## 3rd Qu.: 9.000 Ab : 8 3rd Qu.: 450.0
## Max. :17.000 (Other):25 Max. :1000.0
## NA's :2 NA's : 1
## 'data.frame': 155 obs. of 14 variables:
## $ x : num 181072 181025 181165 181298 181307 ...
## $ y : num 333611 333558 333537 333484 333330 ...
## $ cadmium: num 11.7 8.6 6.5 2.6 2.8 3 3.2 2.8 2.4 1.6 ...
## $ copper : num 85 81 68 81 48 61 31 29 37 24 ...
## $ lead : num 299 277 199 116 117 137 132 150 133 80 ...
## $ zinc : num 1022 1141 640 257 269 ...
## $ elev : num 7.91 6.98 7.8 7.66 7.48 ...
## $ dist : num 0.00136 0.01222 0.10303 0.19009 0.27709 ...
## $ om : num 13.6 14 13 8 8.7 7.8 9.2 9.5 10.6 6.3 ...
## $ ffreq : Factor w/ 3 levels "1","2","3": 1 1 1 1 1 1 1 1 1 1 ...
## $ soil : Factor w/ 3 levels "1","2","3": 1 1 1 2 2 2 2 1 1 2 ...
## $ lime : Factor w/ 2 levels "0","1": 2 2 2 1 1 1 1 1 1 1 ...
## $ landuse: Factor w/ 15 levels "Aa","Ab","Ag",..: 4 4 4 11 4 11 4 2 2 15 ...
## $ dist.m : num 50 30 150 270 380 470 240 120 240 420 ...
Mapping meuse dataset
coordinates(meuse)=~x+y;
data(meuse.riv);
meuse.sr = SpatialPolygons(list(Polygons(
list(Polygon(meuse.riv)),"meuse.riv")))
rv = list("sp.polygons", meuse.sr,
fill = "lightblue")
spplot(meuse["zinc"], do.log = TRUE,
key.space = "bottom",
sp.layout = list(rv),
main = "Zinc Concentration")
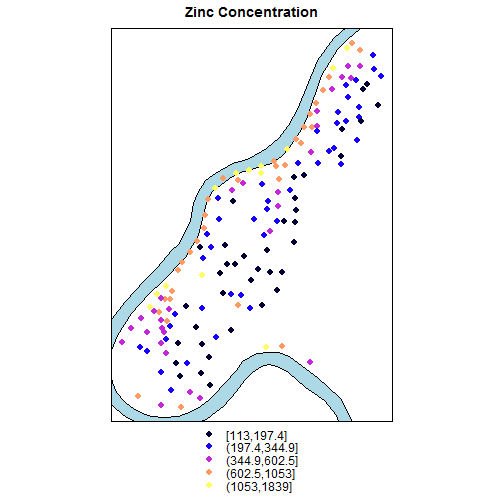
Use spplot to overlay data
data(meuse.grid); coordinates(meuse.grid) = ~x+y;
gridded(meuse.grid) = TRUE
meuse.sl = SpatialLines(list(Lines(list(Line
(meuse.riv)), ID="1")))
par(mar=c(7,0,4,4));
image(meuse.grid["dist"], main = "Relative Magnitude
of Cadmium Concentration & distance from Meuse River");
lines(meuse.sl);
with(meuse, symbols(x=meuse@coords[,1],
y=meuse@coords[,2], circles=meuse@data$cadmium,
inches=1/5, col="black", bg="green", add=T))
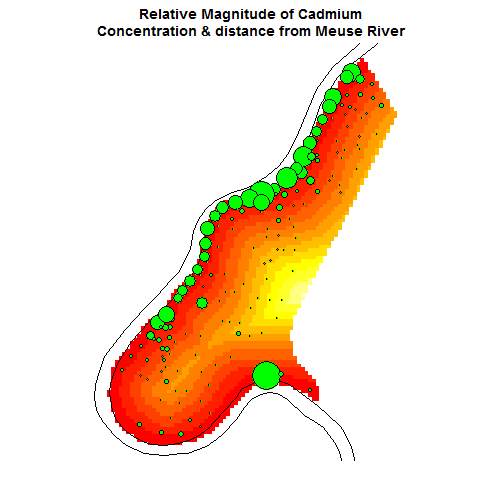
Simple interpolation of zinc concentrations
## [inverse distance weighted interpolation]
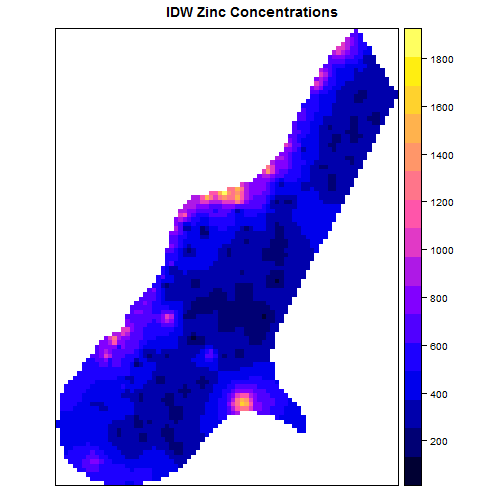
library(gstat)
data(meuse); data(meuse.grid);
coordinates(meuse.grid) = ~x+y;
gridded(meuse.grid) = TRUE
zinc.idw = idw(zinc~1, meuse, meuse.grid)
spplot(zinc.idw["var1.pred"],
main = "IDW Zinc
Concentrations")
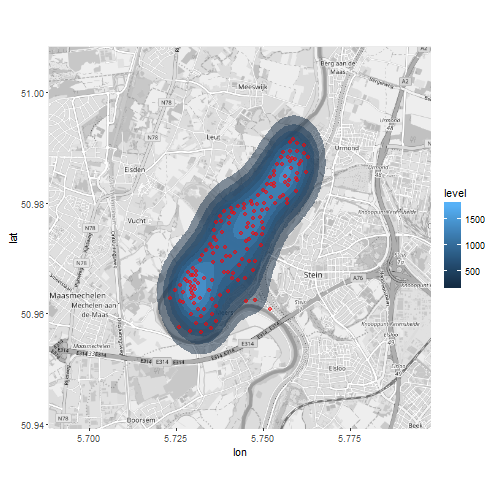
Point density for meuse dataset
library(ggmap); proj4string(meuse) <- CRS("+init=epsg:28992");
meuse_wgs84<-spTransform(meuse, CRS("+init=epsg:4326"))
meuse_wgs84<-data.frame(meuse_wgs84@coords)
meuse_wgs84$copper<-meuse$copper
map <- get_map(c(lon=mean(meuse_wgs84$x), lat=mean(meuse_wgs84$y)), zoom=13, source = "osm",
color = "bw"); mapPoints <- ggmap(map)
mapPoints +
stat_density2d(data=meuse_wgs84,aes(x, y, fill= ..level..), alpha =0.5, geom="polygon") +
geom_point(data=meuse_wgs84,aes(x, y), colour="red", alpha = .5)
Point density for meuse dataset